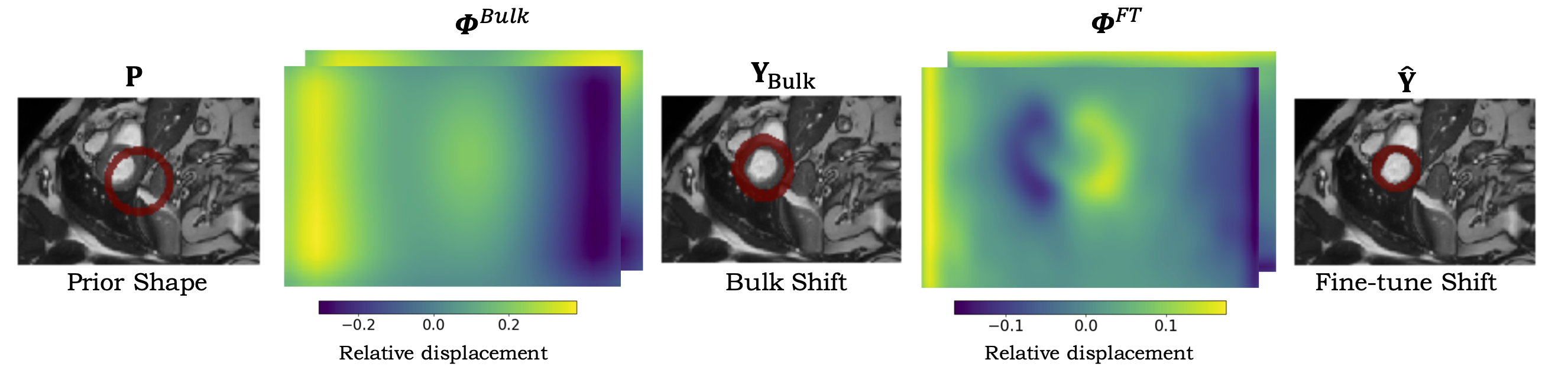
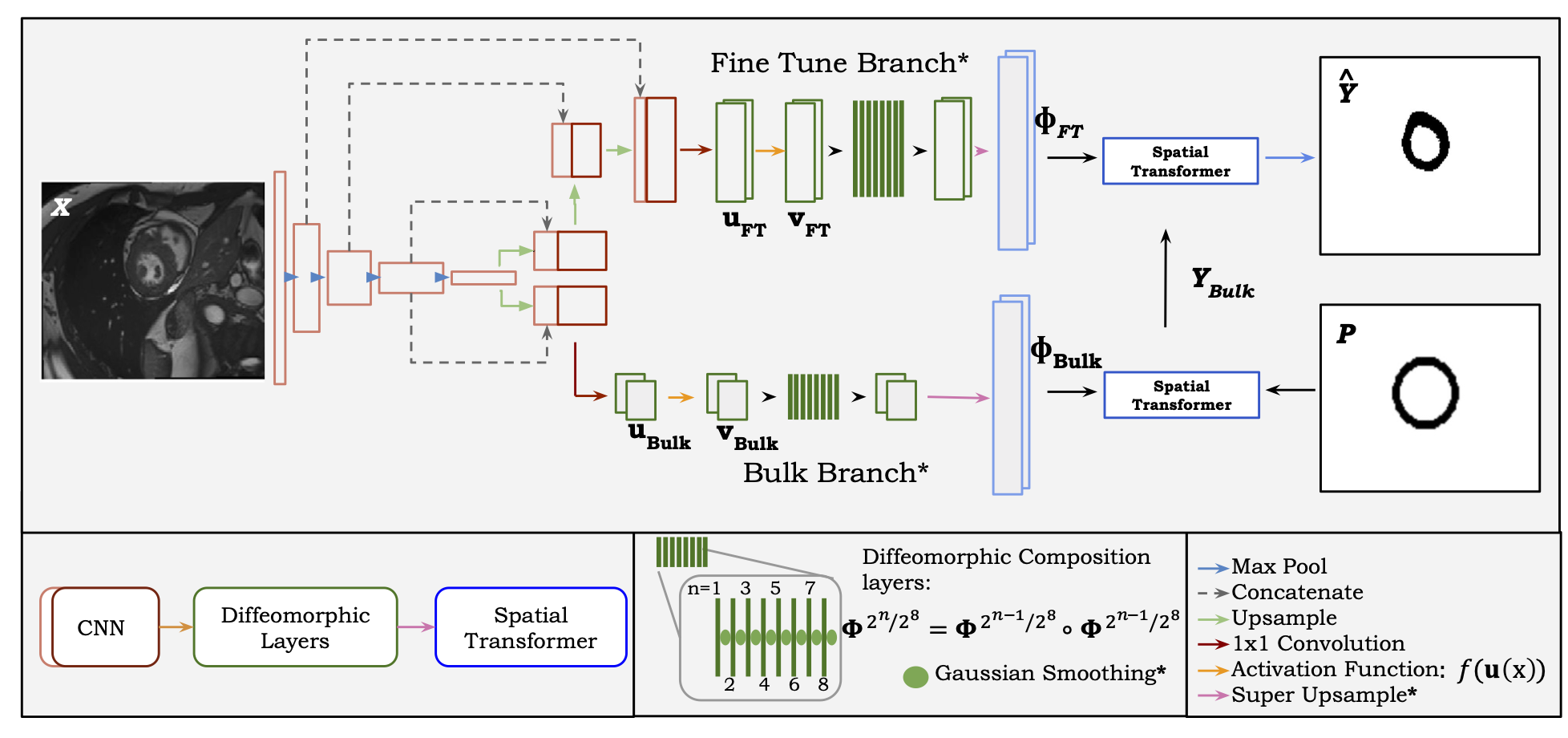
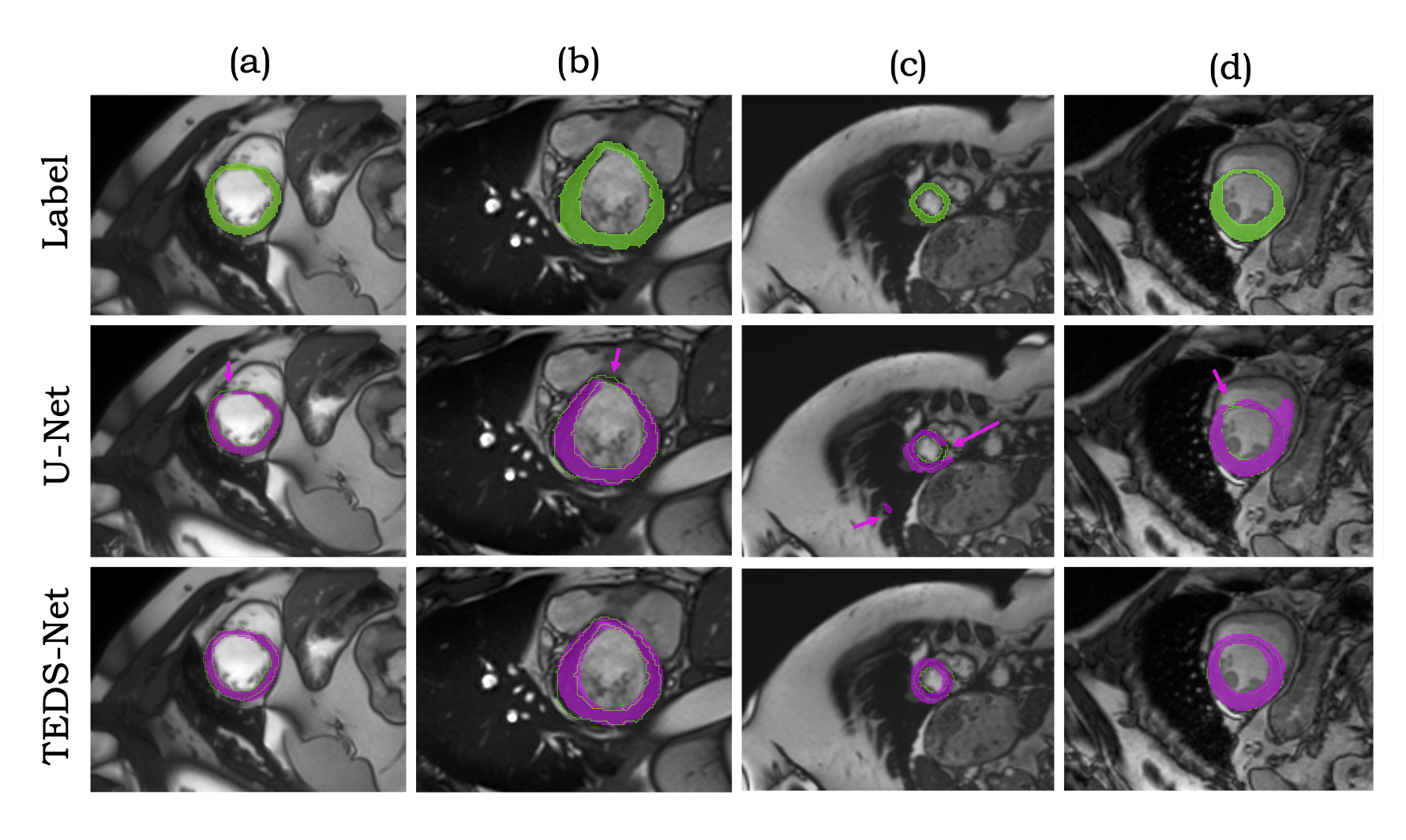
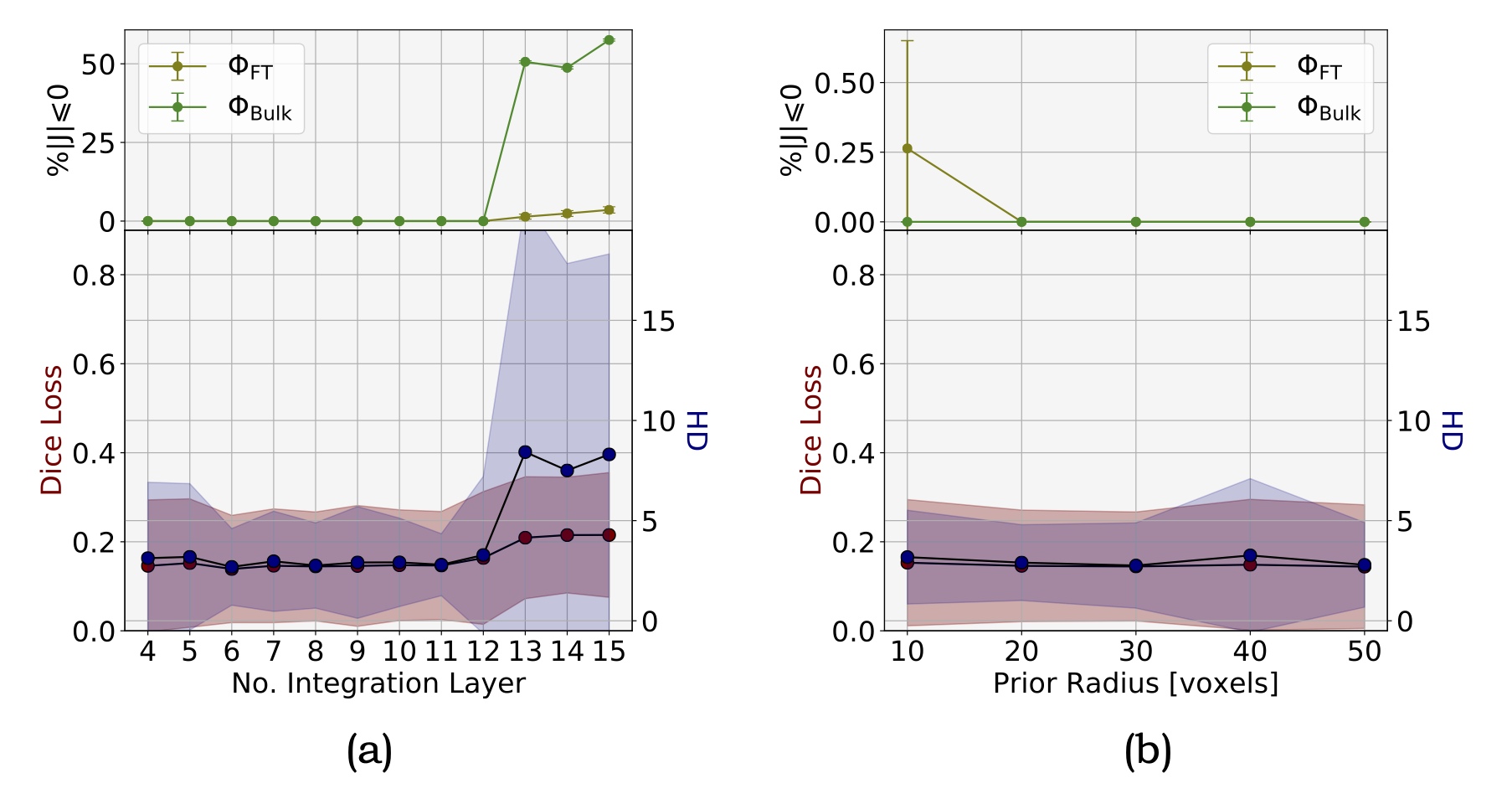
|
Accurate topology is key when performing meaningful anatomical segmentations, however, it is often overlooked in traditional deep learning methods. In this work we propose TEDS-Net: a novel segmentation method that guarantees accurate topology. Our method is built upon a continuous diffeomorphic framework, which enforces topology preservation. However, in practice, diffeomorphic fields are represented using a finite number of parameters and sampled using methods such as linear interpolation, violating the theoretical guarantees. We therefore introduce additional modifications to more strictly enforce it. Our network learns how to warp a binary prior, with the desired topological characteristics, to complete the segmentation task. We tested our method on myocardium segmentation from an open-source 2D heart dataset. TEDS-Net preserved topology in 100% of the cases, compared to 90% from the U-Net, without sacrificing on Hausdorff Distance or Dice performance.
|